Getting started on PySpark on Databricks (examples included)
Gets python examples to start working on your data with Databricks notebooks.


TL;DR;
This article will give you Python examples to manipulate your own data. The example will use the spark library called pySpark.
Prerequisites: a Databricks notebook
-
To get a full working Databricks environment on Microsoft Azure in a couple of minutes and to get the right vocabulary, you can follow this article:
Spark session
Databricks Notebooks have some Apache Spark variables already defined:
-
SparkContext: sc
- Spark Context is an object that tells Spark how and where to access a cluster.
-
SparkSession (Spark 2.x): spark
- Spark Session is the entry point for reading data and execute SQL queries over data and getting the results.
- Spark session is the entry point for SQLContext and HiveContext to use the DataFrame API (sqlContext).
All our examples here are designed for a Cluster with python 3.x as a default language. That's why we do not need the magic keyword %python on the first line. This is however required for Scala cells of our notebook.
print("Spark version", sc.version, spark.sparkContext.version, spark.version)
print("Python version", sc.pythonVer)- Same in Scala:
%scala
println("Spark version " + sc.version)
println("Spark version " + spark.version)
println("Spark version " + spark.sparkContext.version)
println("Scala version " + scala.util.Properties.versionString)RDD (schema-less) vs DF
RDD: Resilient Distributed Dataset (RDD)
An RDD is an immutable distributed collection of data partitioned across nodes in your cluster with a low-level API. It is schema-less and used for low-level transformation and actions.
Dataframe (DF)
A DataFrame is a distributed collection of rows under named columns. It is the same as a table in a relational database. It is closed to Pandas DataFrames. A DataFrame has the ability to handle petabytes of data and is built on top of RDDs. A DataFrame is mapped to a relational schema.
Create a RDD
myRDD = sc.parallelize(
[('Amber', 22), ('Alfred', 23), ('Skye',4), ('Albert', 12),
('Amber', 9)])Display the content of the RDD
# Option 1
print(myRDD.collect())
# Option 2
for row in myRDD.take(myRDD.count()): print(row)Convert a RDD to a DF
- You need to give the schema to use:
from pyspark.sql import types
DFSchema = types.StructType([
types.StructField('name', types.StringType()),
types.StructField('age', types.LongType())
])
# 3 ways to convert the RDD to a DF:
df1 = spark.createDataFrame(myRDD, DFSchema)
df2 = myRDD.toDF(DFSchema)
df3 = myRDD.toDF(['name','age'])Display the dataframe content (first 1000 rows)
display(df1)Display the DF schema
df1.printSchema()The number of rows
df1.count()The column names
df1.columnsThe number of partitions
The dataframe is divided in partitions. Each time an evaluation is requested, a job is created and the DF is distributed among workers on the cluster. The entity that is distributed to each worker is a partition. The number of partitions is defined automatically and can also be set by hand. It is recommended to adjust the number of partitions especially to reduce it if you do have a very small dataframe. In that case, the data distribution will add an overhead.
# Get the partition number
print("Partitions", df1.rdd.getNumPartitions())
# Set the number of partition (re-repartition)
df1 = df1.repartition(2)Alias the DF in the SQL context
df1.createOrReplaceTempView("user")Perform SQL queries
- Embedded in Python
result_df = spark.sql("SELECT * from user")- In a SQL cell
%sql
SELECT * from userExamples of DF queries
display(df1.select("name", "age").where("name = 'Amber'"))display(df1.select("name", "age").where("name = 'Amber'"))from pyspark.sql import functions as F
display(df1.select("*").where(F.col("name").eqNullSafe("Amber")))display(df1.select("name", "age").where(F.col("name").eqNullSafe("Amber")))display(df1.select(*df1.columns).where(F.col("name").eqNullSafe("Amber")))display(df1.select(F.min("age").alias("minValue"), F.max("age")))Regular python interactions
- Get the DF in regular python list
pythonRegularObject = df1.collect()- Destructuring the result in regular Python variables
minValue, maxValue, = df1.select(F.min("age"), F.max("age")).head()
print("Min: {}, Max: {}".format(minValue, maxValue))- Same in Scala
%scala
import org.apache.spark.sql.functions._
var df = spark.table("user")
// df = spark.sql("select * from user")
var Row(minValue, maxValue) = df.select(min("age"), max("age")).head
println(s"Min: ${minValue}, Max: ${maxValue}")User Defined Functions (UDF)
- Without UDF
result_df = df1.select("name", "age", (F.col("age") * 2).alias("ageX2"))
display(result_df)- With an UDF
@udf("long")
def multiplyBy2(age):
return age * 2
result_df = df1.select("name", "age", multiplyBy2("age").alias("ageX2"))
display(result_df)Important note: avoid UDF as much as you can as they are slow (especially in Python) compared to native pySpark functions.
DF to RDD and vise versa (map, flatmap)
- Applying a python function on each row thanks to the RDD function
mapto create a new columnageX2:
from pyspark.sql import Row
def tranformRow(row):
newAge = row.age * 2
return Row(name=row.name, age=row.age, ageX2=newAge)
# return Row(**{"name": row.name, "age": row.age, "ageX2": newAge})
result_df = df1.rdd.map(tranformRow).toDF()
display(result_df)- Advanced example using Python generator (
yieldkeyword) andflatmapto create new rows:
# Advanced
def rowExpander(x):
yield Row(**{"name": x.name, "age": x.age})
yield Row(**{"name": x.name, "age": x.age * 2})
result_df = df1.rdd.flatMap(rowExpander).toDF()
display(result_df)- RDD api can also be used to get back regular python array:
df1.select("name").distinct().rdd.flatMap(lambda x: x).collect()File handling
Magic %fs
- Use the %fs magic keyword:
%fs
ls /mntLoading a JSON file
- Load a Json file (the schema will be inferred)
# Option 1
df = spark.read.format("json").load("dbfs:/mnt/my_file.json")
# Option 2
df = spark.read.json("dbfs:/mnt/my_file.json")- You can speed up the JSON loading by providing the schema:
from pyspark.sql.types import *
dfSchema = StructType([
StructField("id",LongType(),True),
StructField("timestamp",LongType(),True),
StructField("message",StringType(),True),
StructField("signals",ArrayType(StructType(
[
StructField("key",StringType(),True),
StructField("value",DoubleType(),True)
]),True),True)
])
# Providing schema speed up the loading
df = spark.read.format("json").schema(dfSchema).load("dbfs:/mnt/my_file.json")
display(df.limit(5))- A lot of different file formats are supported (CSV, JSON, Parquet, ORC, etc...).
DF handling examples
Explode
- If you have a JSON file where each JSON is a row like this:
{
"id": 1,
"timestamp": 0001,
"message": "message1",
"signals": {
"prop1": "value1",
"prop2": "value2",
}
}
{
"id": 2,
"timestamp": 0002,
"message": "message2",
"signals": {
"prop3": "value3",
"prop4": "value4",
}
}You can use the explode function to convert it into a DF like this:
id | timestamp | message | key | value
1 | 0001 | message1 | prop1 | value1
1 | 0001 | message1 | prop2 | value2
2 | 0002 | message2 | prop3 | value3
2 | 0002 | message2 | prop4 | value4 dfExploded = (
df.select("id", "timestamp", "message", F.explode("signals").alias("signal"))
.select("id", "timestamp", "message", F.col("signal.key").alias("key"), F.col("signal.value").alias("value"))
)
display(dfExploded.limit(5))display(dfExploded.filter("message='message1'"))group by
ret = (
dfExploded
.filter("message='message1'")
.groupBy("key")
.agg(F.mean("value"))
)
display(ret)Cache result to speed up further processing
intermediateDF = dfExploded.filter("message='message1'").cache()Pivot
ret = (
intermediateDF
.groupBy("timestamp")
.pivot("key", ["prop1", "prop2"])
.agg(F.first("value"))
)
display(ret)Plotting
There is some built-in plotting with Databricks. However if you want more control, you can use external library like plotly](https://plot.ly/).
- Getting some data to plot:
import requests
r = requests.get("https://timeseries.surge.sh/usd_to_eur.csv")
df = spark.read.csv(sc.parallelize(r.text.splitlines()), header=True, inferSchema=True)
display(df)- Using plotly
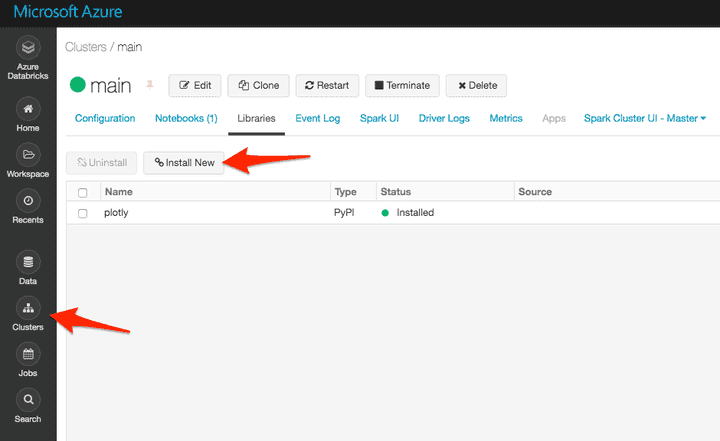
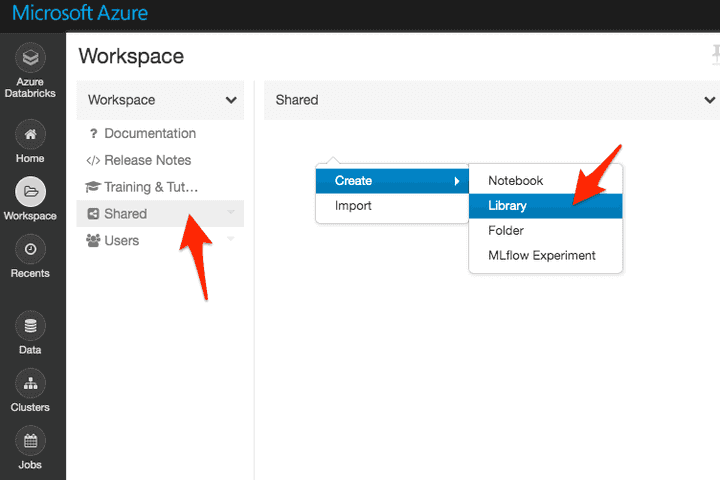
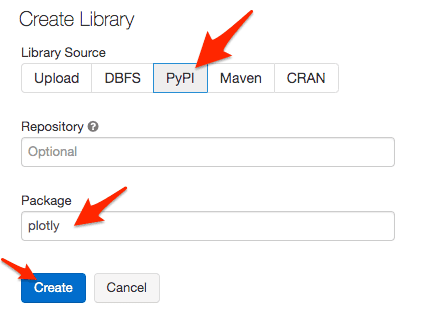
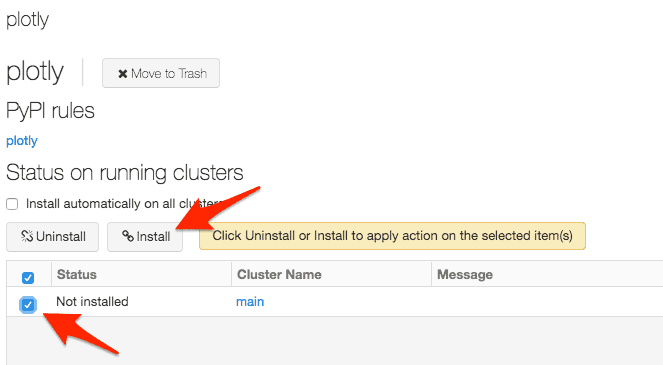
Important note: you need to add plotly as a cluster dependency:
Option 1
Option 2
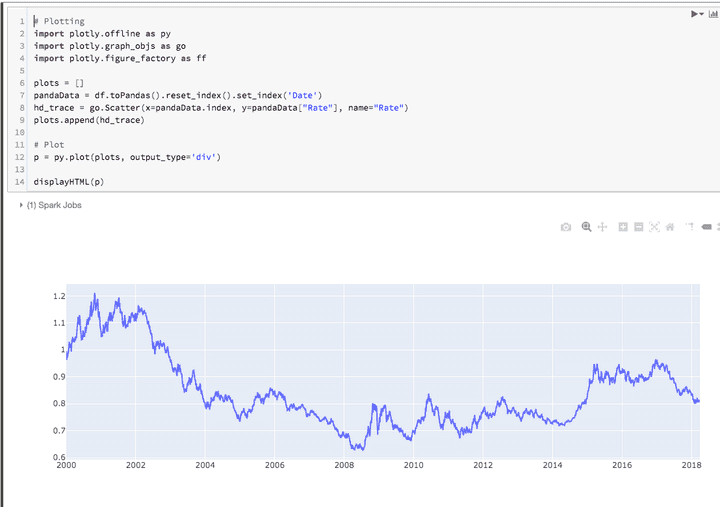
# Plotting
import plotly.offline as py
import plotly.graph_objs as go
import plotly.figure_factory as ff
plots = []
pandaData = df.toPandas().reset_index().set_index('Date')
hd_trace = go.Scatter(x=pandaData.index, y=pandaData["Rate"], name="Rate")
plots.append(hd_trace)
# Plot
p = py.plot(plots, output_type='div')
displayHTML(p)Windows
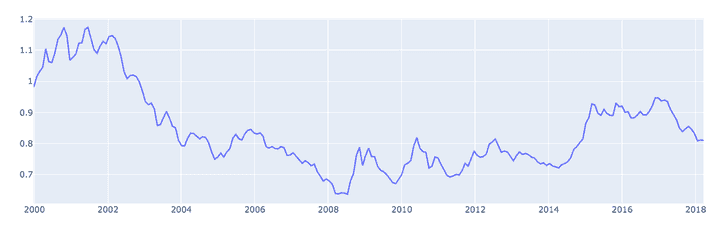
- We will create a tumbling window to downsample the data to 1 point every 30 days.
- We will use the
meanfunction.
tumblingSize = "30 days"
downsampledDf = (
df
.groupBy(F.window("Date", tumblingSize).alias("timestamp"))
.agg(F.mean("Rate").alias("Rate"))
.select(F.col("timestamp.start").alias("Date"), "Rate")
.sort(F.asc("Date"))
)
display(downsampledDf)- Plot again:
plots = []
pandaData = downsampledDf.toPandas().reset_index().set_index('Date')
hd_trace = go.Scatter(x=pandaData.index, y=pandaData["Rate"], name="Rate")
plots.append(hd_trace)
# Plot
p = py.plot(plots, output_type='div')
displayHTML(p)👉 Don't forget to follow me on Twitter to be notified when new posts are available!